Resources
mergem: merging, comparing, and translating genome-scale metabolic models
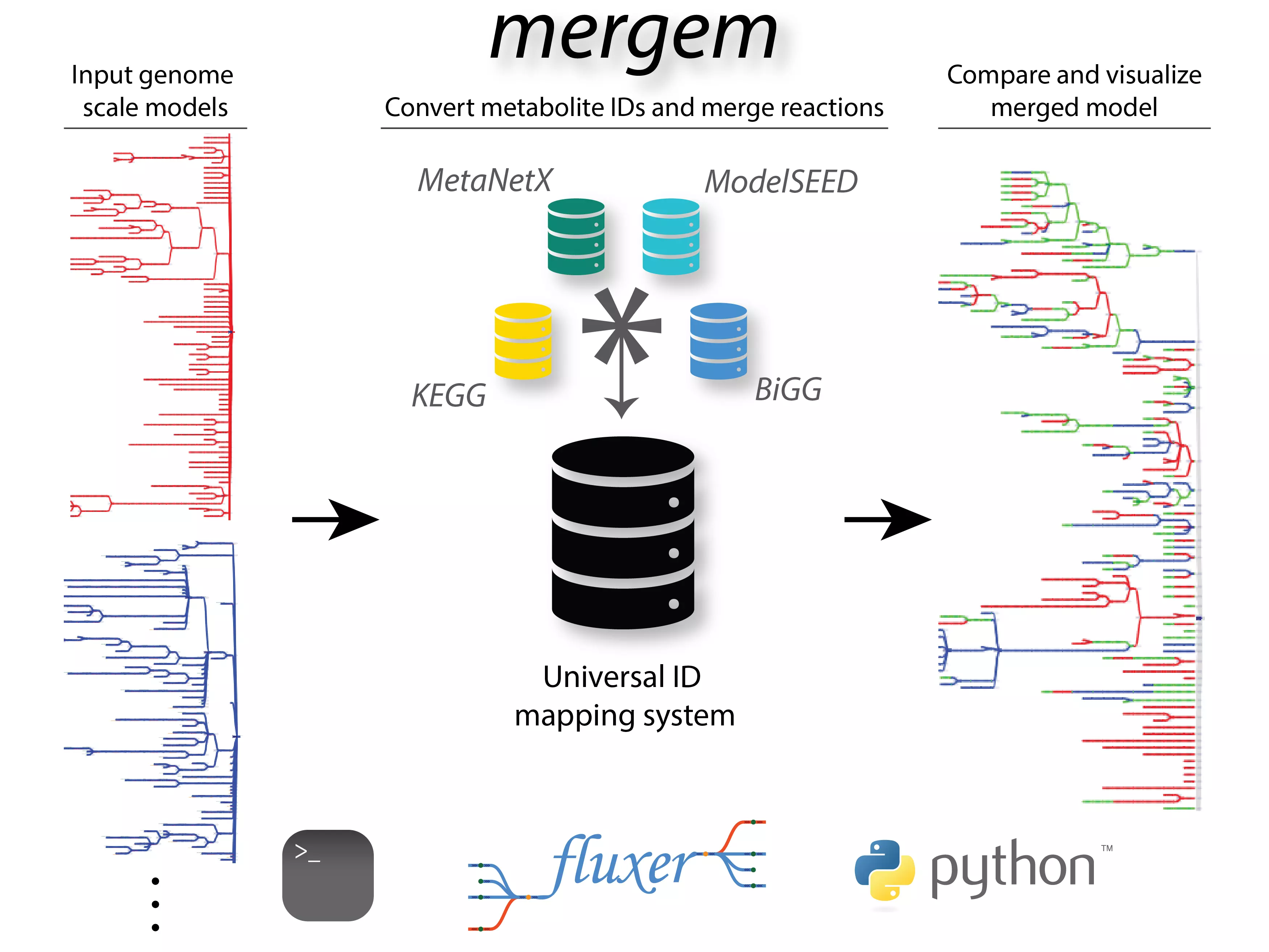
mergem is a Python package and command-line tool for comparing and merging genome-scale metabolic models and draft reconstructions. The tool translates and unifies metabolite identifiers using a comprehensive database mapping system and then compares reactions based on their participating reactant and product metabolites. The method is freely available as a Python package and command line tool. In addition, mergem has been integrated into our user-friendly Fluxer web application, which allows any user without programming knowledge to perform the merging, comparison, and visualization of any number of models directly in the web application.
Streamlined gene cloning with Gibson assembly
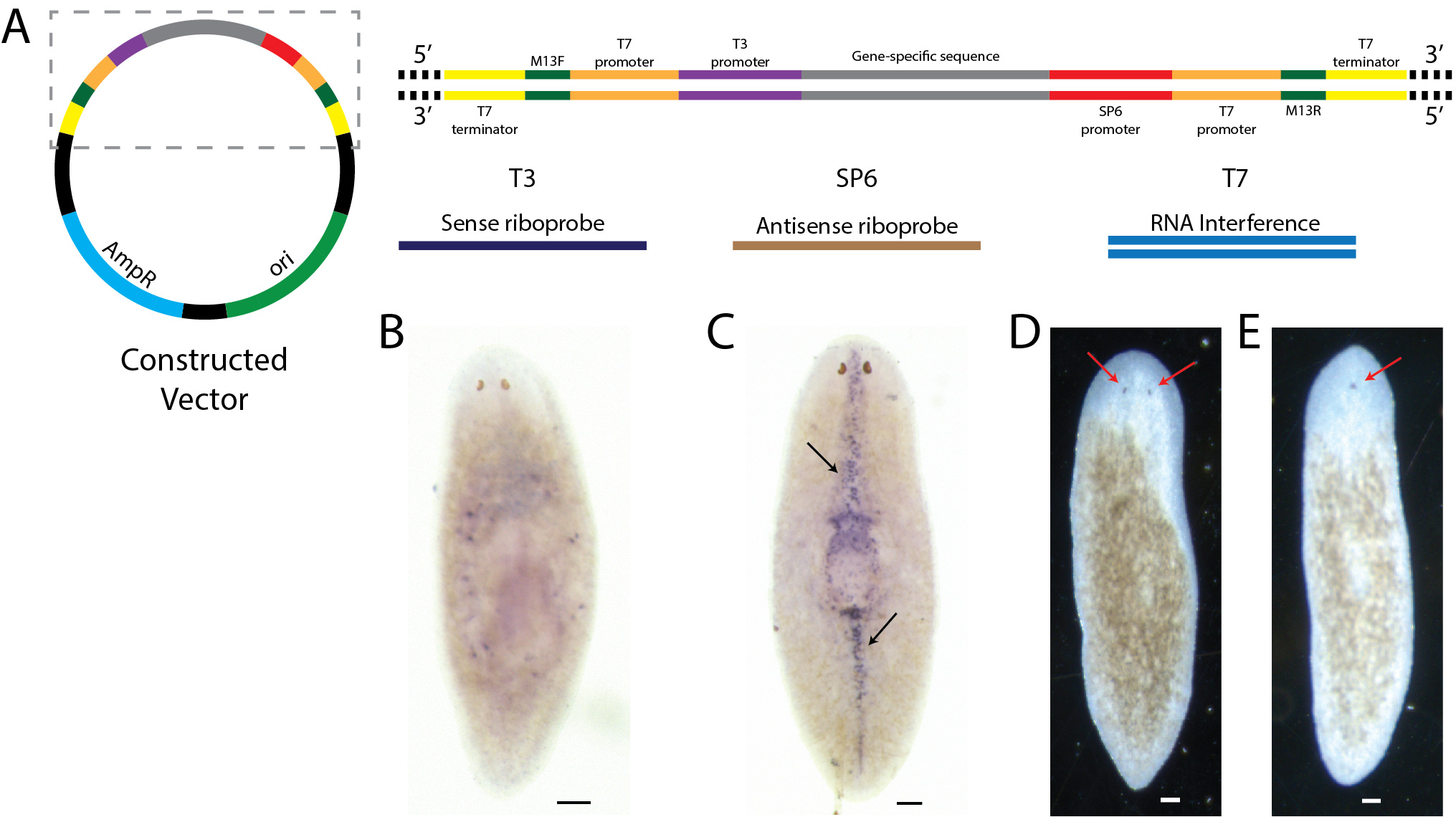
We have developed a cloning vector and a protocol for a faster gene cloning with Gibson assembly. The protocol allows the simultaneous plasmid restriction digestion and Gibson assembly, saving time in the cloning of genes. The novel cloning vector is ideal for the synthesis of both riboprobes and double-stranded RNAs for in situ and RNA interference experiments, respectively. We have illustrated the protocol with planaria in situ and RNAi assays, but it is applicable to any organism.
AutoPortrait: automatic generation of interactive multidimensional phase portraits
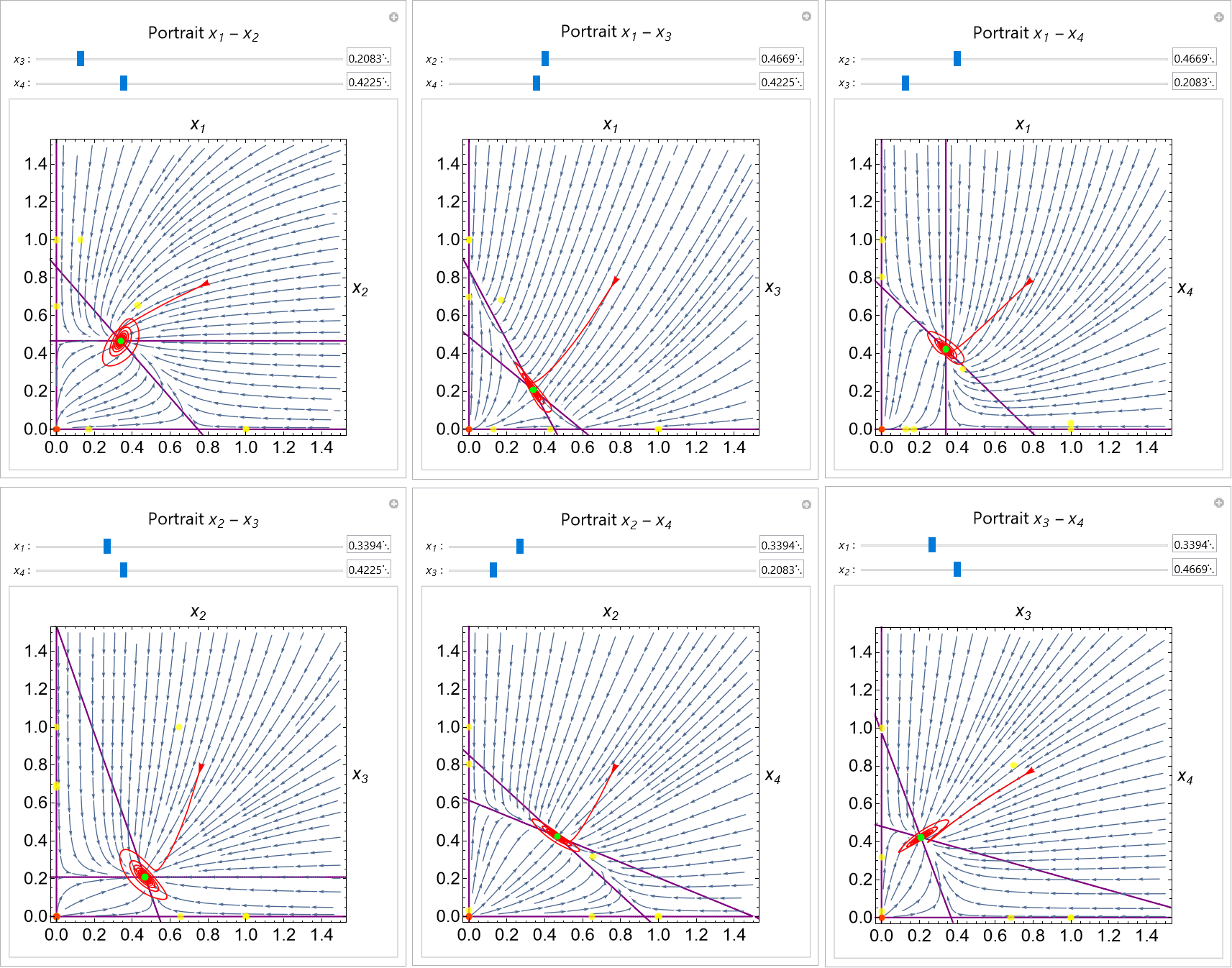
Autoportrait is a Mathematica package and user-friendly interface for the automatic generation of interactive phase portraits for dynamical systems. The portraits generated are interactive and the user can move the visualized planar slice of the domain, change parameters with sliders, and add trajectories in the phase and time domains. The diagrams are updated in real time and can be saved in computable notebooks. The method can be accessed with a user-friendly interface or called directly from scripts.
Fluxer: flux and pathways viewer
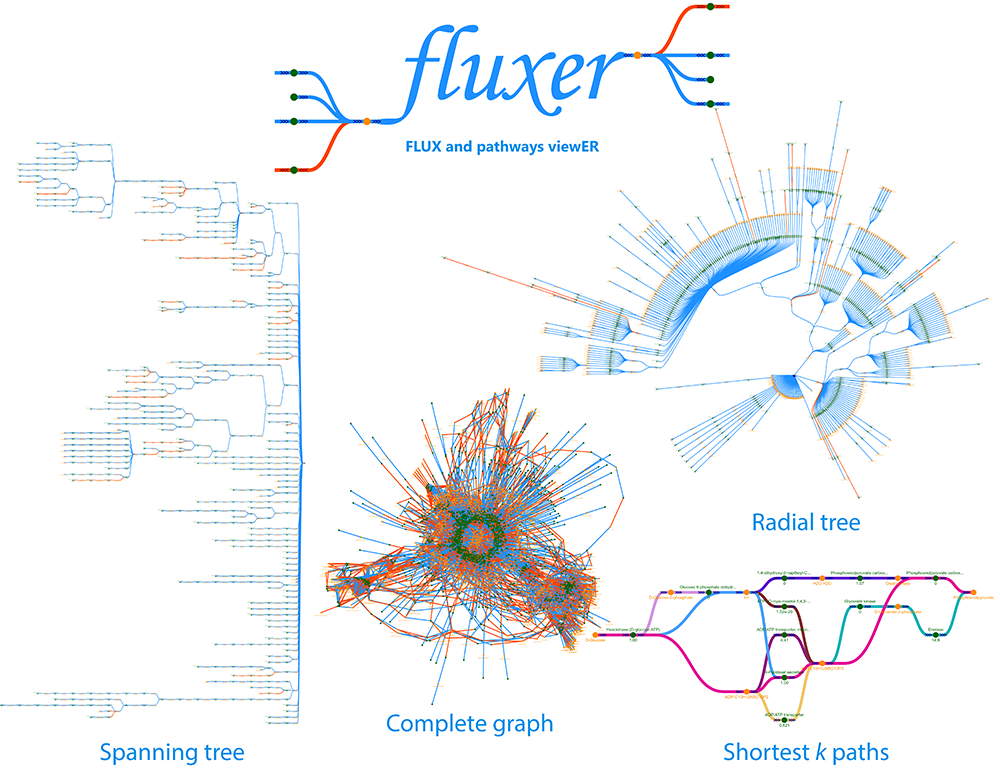
Fluxer is a web application for the computation and interactive visualization of flux networks from genome-scale metabolic models. Fluxer can perform flux balance analysis and visualize the resultant flux networks with spanning trees, k-shortest paths, and complete graphs. More than 80 genome-scale models are currently available in the server, and any user can upload any SBML model to analyze it with the tool. Fluxer can simulate enzymatic knock-outs and calculate their effects in terms of flux and growth. Models with thousands of reactions and metabolites can be efficiently visualized and analyzed with Fluxer.
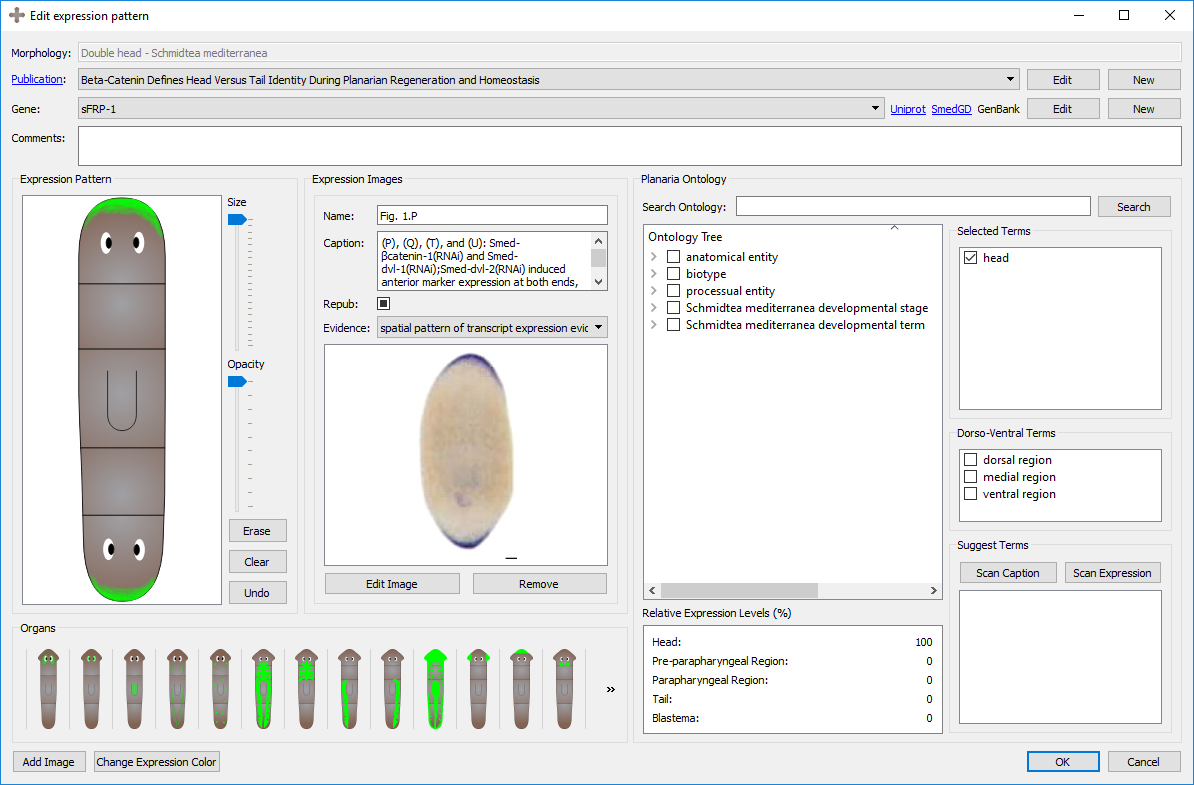
PlanGexQ: planarian gene expression patterns curation tool
PlanGexQ is a user-friendly application for the curation and ontological annotation of planarian morphologies and gene expression patterns. Reference worm morphologies can be defined with mathematical graphs using a drag-and-drop interface, and microscopy images of gene expression patterns can be registered into them through the user interface. The program can suggest ontological gene annotations based on the gene expression patterns and their textual descriptions, which are stored in a centralized database together with the images, registrations, and annotated descriptions.
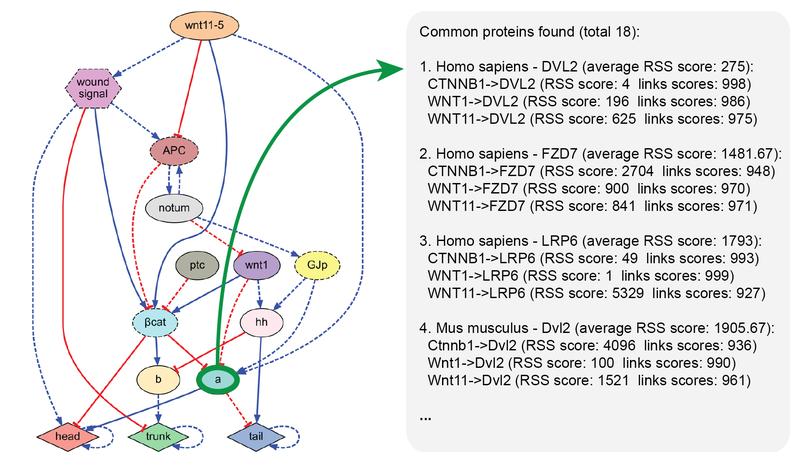
MoCha: molecular characterization
MoCha is a software tool for finding unknown proteins and their pathways interacting with a given set of proteins. Due to its highly optimized pre-processing, MoCha can search within seconds through more than a billion interactions from over 2,000 organisms in the STRING database. This tool is useful for working on network models by hand or as an integrated software pipeline and can readily aid in the reverse-engineering of regulatory models.
Planform: planarian formalization
Planform is a database and user-friendly application for the design, storage, and search of formally-encoded planarian regenerative experiments, manipulations, and morphologies. It contains more than 1,000 different experiments curated from the literature and provides a graphical user interface that allows any scientist to query, extend, and search in this large dataset.
Limbform: limb formalization
Limbform is a database and software tool that centralizes with a mathematical ontology the details of more than 800 experiments of limb regeneration curated from the literature and comprising many model organisms, including salamanders, frogs, insects, crustaceans and arachnids. The user-friendly software tool can be used for mining the curated experiments, as well as for formalizing new ones.
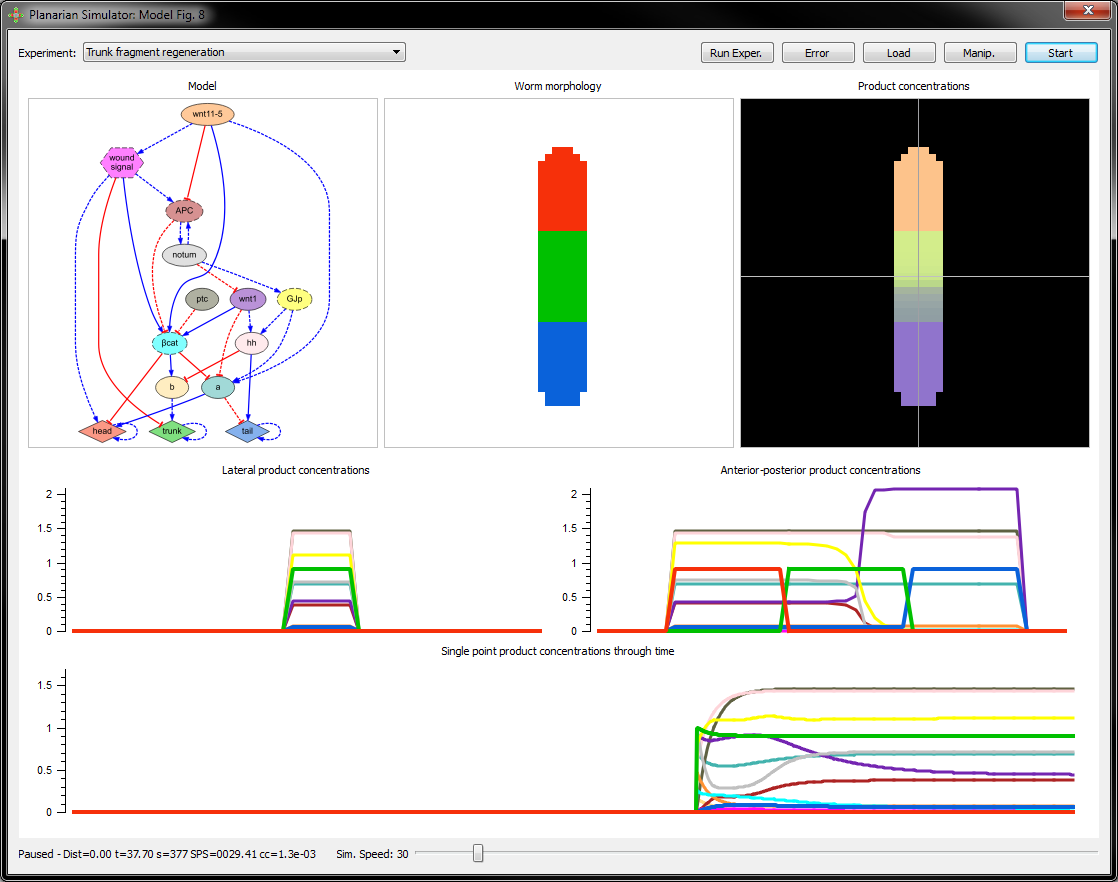
Planarian dynamic models
We have created a computational method that can reverse-engineer dynamic regulatory networks directly from experimental results. A software application is available to visualize and interact with the discovered models of planarian regeneration. You can download the application and simulate planarian regeneration experiments with the inferred models in your own computer.