Systems Biology
Dynamic signaling networks
We are interested in computational and mathematical models based on the theory of computer science and dynamical systems to describe, simulate, and predict the behavior of complex biological processes. We study dynamic models and perform in-silico experiments involving form, shape, and patterning formation, signaling mechanisms responsible for cell behaviors, and synthetic regulatory networks engineered to perform specific functions. These models aid us to predict the outcomes of novel perturbations, understand why these regulatory mechanisms can go awry such as in cancer, and, more importantly, find interventions that can repair and restore their original functionality.
Pattern, shape, and form regulation
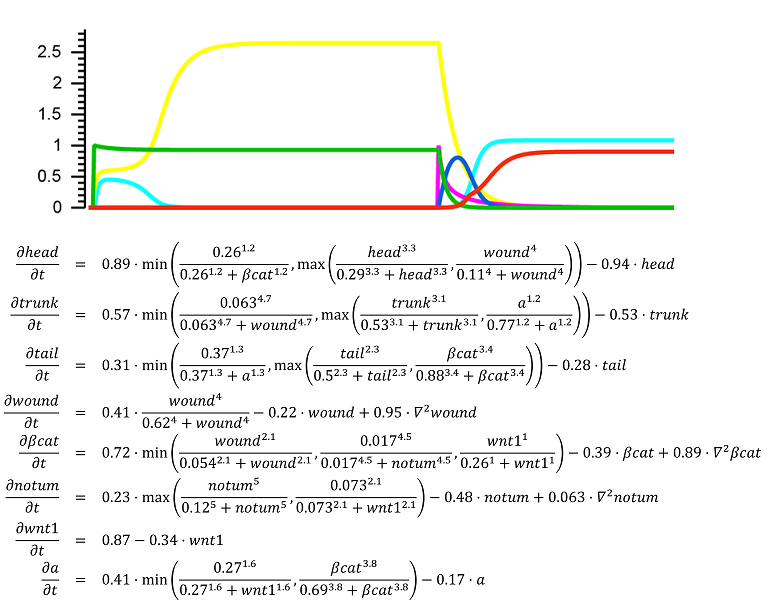
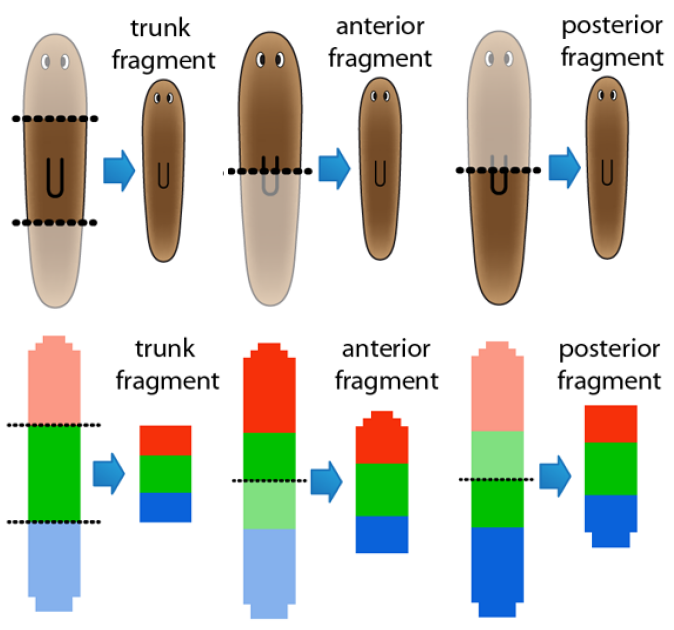
The regulation of the formation of biological patterns, shapes, and forms has been historically one of the most difficult aspects to understand in biology. The complexity of biological regulation together with the multi-dimensional spatial characteristics of these processes represent a hard barrier for our comprehension. We seek to discover mechanistic models to aid us to understand, predict, and control the formation of biological spatial properties. We use dynamic models at the organism and tissue levels to simulate patterns, shapes, and forms together with the signaling mechanisms controlling their formation. These spatial dynamic models are already paving the way for the long sought comprehensive theory of growth and form.
Stochastic outcomes
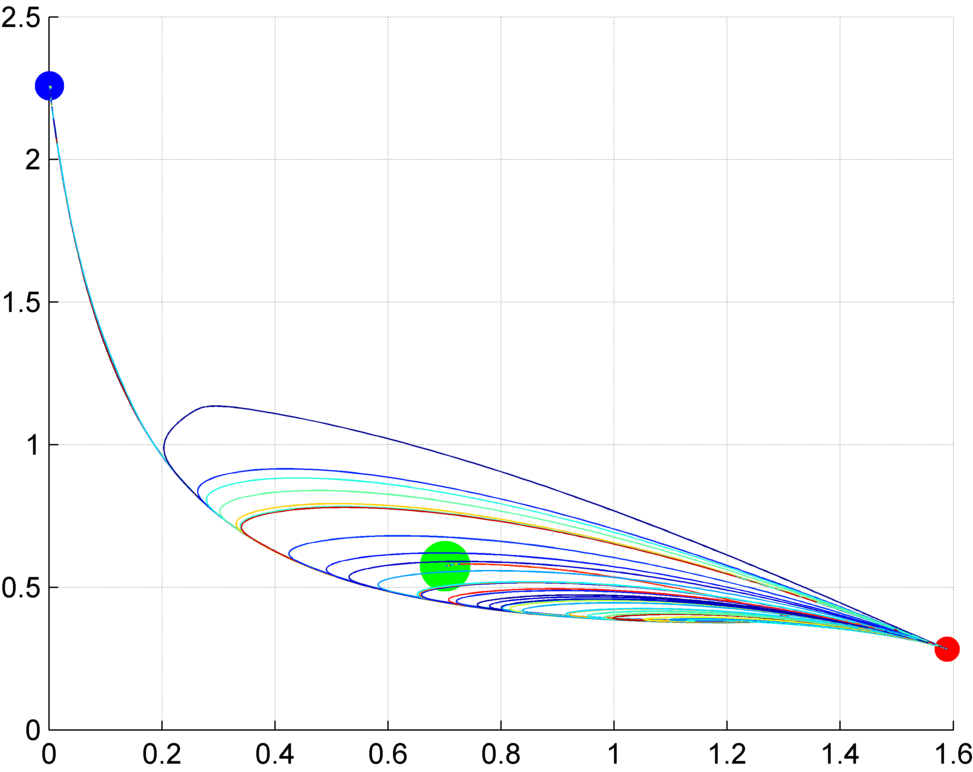
Interestingly, most biological experiments result in stochastic outcomes. Knocking down certain genes or applying a pharmacological drug to genetically-clone animals usually produce a set of different results, each of them with a specific probability. Is this stochasticity due to the noise of biological interactions, some unknown variability within the organisms, or an inherent property of the signaling mechanisms? We investigate stochastic dynamic models to discover the source of this variability, study their dynamic properties, such as their space state and dynamic attractors, and how specific pharmacological and genetic interventions can create bifurcations in the system. Importantly, these analyses can reveal possible treatments to revert undesirable conditions characteristic of certain diseases.
Publications
Discovery of dynamic models for AML disease progression from longitudinal multi-modal clinical data using explainable machine learning
R. Mousavi, M.K. Mustafa Ali, D. Lobo
medRxiv doi:10.1101/2025.04.07.25325267, 2025.
Emergent tissue shapes from the regulatory feedback between morphogens and cell growth
B. Kaity, D. Lobo
bioRxiv doi:10.1101/2025.02.16.638504, 2025.
FROG analysis ensures the reproducibility of genome scale metabolic models
K. Raman, M. Kratochvil, et al., D. Lobo, et al., R.S. Malik-Sheriff
bioRxiv doi:10.1101/2024.09.24.614797, 2024.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, A. Wolff, D. Lobo
Development 151 (9): dev202353, 2024.
(Highlighted and interviewed)
Automatic design of gene regulatory mechanisms for spatial pattern formation
R. Mousavi, D. Lobo
NPJ Systems Biology and Applications 10, 35, 2024.
mergem: merging, comparing, and translating genome-scale metabolic models using universal identifiers
A. Hari, A. Zarrabi, D. Lobo
NAR Genomics and Bioinformatics 6(1), lqae010, 2024.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, D. Lobo
bioRxiv doi:10.1101/2023.09.15.557968, 2023.
mergem: merging and comparing genome-scale metabolic models using universal identifiers
A. Hari, D. Lobo
bioRxiv doi:10.1101/2022.07.14.499633, 2022.
In situ probe and inhibitory RNA synthesis using streamlined gene cloning with Gibson assembly
A. Wolff, C. Wagner, J. Wolf, D. Lobo
STAR Protocols 3, 101458, 2022.
Formalizing phenotypes of regeneration
D. Lobo
Whole-Body Regeneration
S. Blanchoud, B. Galliot (eds.)
Springer pp. 663-679, 2022.
Automatic generation of interactive multidimensional phase portraits
O.O. Ogunsan, D. Lobo
bioRxiv doi:10.1101/2022.02.23.481676, 2022.
Inference of dynamic spatial GRN models with multi-GPU evolutionary computation
R. Mousavi, S.H. Konuru, D. Lobo
Briefings in Bioinformatics 22(5), bbab104, 2021.
Computational systems biology of morphogenesis
J.M. Ko, R. Mousavi, D. Lobo
Computational Systems Biology in Medicine and Biotechnology
S. Cortassa, M.A. Aon (eds.)
Springer pp. 343-365, 2021.
Kinetic modeling of microbial growth, enzyme activity, and gene deletions: an integrated model of β-glucosidase function in Cellvibrio japonicus
J. Hwang, A. Hari, R. Cheng, J.G. Gardner, D. Lobo
Biotechnology and Bioengineering 117, pp. 3876-3890, 2020.
Fluxer: a web application to compute, analyze, and visualize genome-scale metabolic flux networks
A. Hari, D. Lobo
Nucleic Acids Research 48, pp. 427-435, 2020.
Continuous dynamic modeling of regulated cell adhesion: sorting, intercalation, and involution
J.M. Ko, D. Lobo
Biophysical Journal 117(11), pp. 2166-2179, 2019.
Cross-inhibition of Turing patterns explains the self-organized regulatory mechanism of planarian fission
S. Herath, D. Lobo
Journal of Theoretical Biology 485, 110042, 2019.
Continuous dynamic modeling of regulated cell adhesion
J.M. Ko, D. Lobo
bioRxiv 582429, 2019.
Modeling regenerative processes with Membrane Computing
M. García-Quismondo, M. Levin, D. Lobo
Information Sciences 381, pp. 229-249, 2017.
Discovering novel phenotypes with automatically inferred dynamic models: a partial melanocyte conversion in Xenopus
D. Lobo, M. Lobikin, M. Levin
Scientific Reports 7, 41339, 2017.
Computing a worm: reverse-engineering planarian regeneration
D. Lobo, M. Levin
Advances in Unconventional Computing: Emergence, Complexity and Computation
A. Adamatzky (ed.)
Springer pp. 637-654, 2017.
Computational discovery and in vivo validation of hnf4 as a regulatory gene in planarian regeneration
D. Lobo, J. Morokuma, M. Levin
Bioinformatics 32(17), pp. 2681-2685, 2016.
Physiological controls of large-scale patterning in planarian regeneration: a molecular and computational perspective on growth and form
F. Durant, D. Lobo, J. Hammelman, M. Levin
Regeneration 3(2), pp. 78-102, 2016.
(Selected for the journal cover)
MoCha: molecular characterization of unknown pathways
D. Lobo, J. Hammelman, M. Levin
Journal of Computational Biology 23(4): 291-297, 2016.
A dynamic architecture of life
B.P. Rubin, J. Brockes, B. Galliot, U. Grossniklaus, D. Lobo, M. Mainardi, M. Mirouze, A. Prochiantz, A. Steger
F1000Research 4:1288, 2015.
Serotonergic regulation of melanocyte conversion: A bioelectrically regulated network for stochastic all-or-none hyperpigmentation
M. Lobikin, D. Lobo, D.J. Blackiston, C.J. Martyniuk, E. Tkachenko, M. Levin
Science Signaling 8(397), pp. ra99, 2015.
(Reviewed in a focus paper)
Inferring regulatory networks from experimental morphological phenotypes: a computational method reverse-engineers planarian regeneration
D. Lobo, M. Levin
PLoS Computational Biology 11(6): e1004295, 2015.
A linear-encoding model explains the variability of the target morphology in regeneration
D. Lobo, M. Solano, G.A. Bubenik, M. Levin
Journal of the Royal Society Interface 30(24), pp. 3598-3600, 2014.
(Recommended by F1000Prime, Faculty of 1000, 718232471)
Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo
M. Lobikin, B. Chernet, D. Lobo, M. Levin
Physical Biology 9(6): 065002, 2012.
(Selected for the journal cover)
Modeling planarian regeneration: a primer for reverse-engineering the worm
D. Lobo, W.S. Beane, M. Levin
PLoS Computational Biology 8(4): e1002481, 2012.
(Selected for the journal cover)