Ontologies and Databases
Formalisms and ontologies
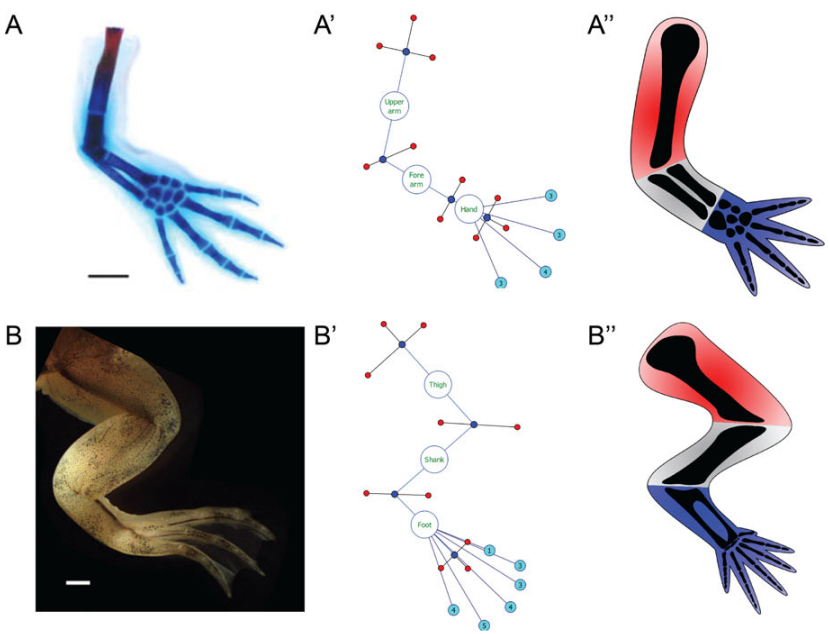
Natural language (such as English) is inherently ambiguous and can contain contradictions and omit details, yet it is commonly used to disseminate methods, experiments, and results in scientific papers. Moreover, natural language represents a barrier for applying automated computational methods to the vast amount of scientific information available in the literature. We investigate formalisms and ontologies based on mathematical language to unambiguously describe complex experimental procedures and their morphological outcomes. Using mathematical descriptions is not only necessary for semantic clarity for human scientists, but a requirement for the application of artificial intelligence systems able to understand the data.
Databases
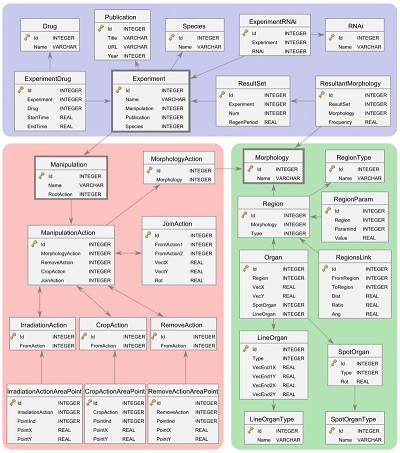
Based on mathematical formalisms and ontologies, we develop and curate databases that centralize the experimental knowledge disseminated in the literature. These databases represent an extraordinary resource for the community, paving the way for the streamlining of scientific data. In addition, the mathematical nature of the formalisms and ontologies that these databases are built with allows the direct application of artificial intelligence systems to access and understand the data. Indeed, our curated databases represent the first step towards the application of automated systems that can extract new knowledge and reverse-engineer mechanistic models from the outstanding results obtained at the bench.
Expert systems
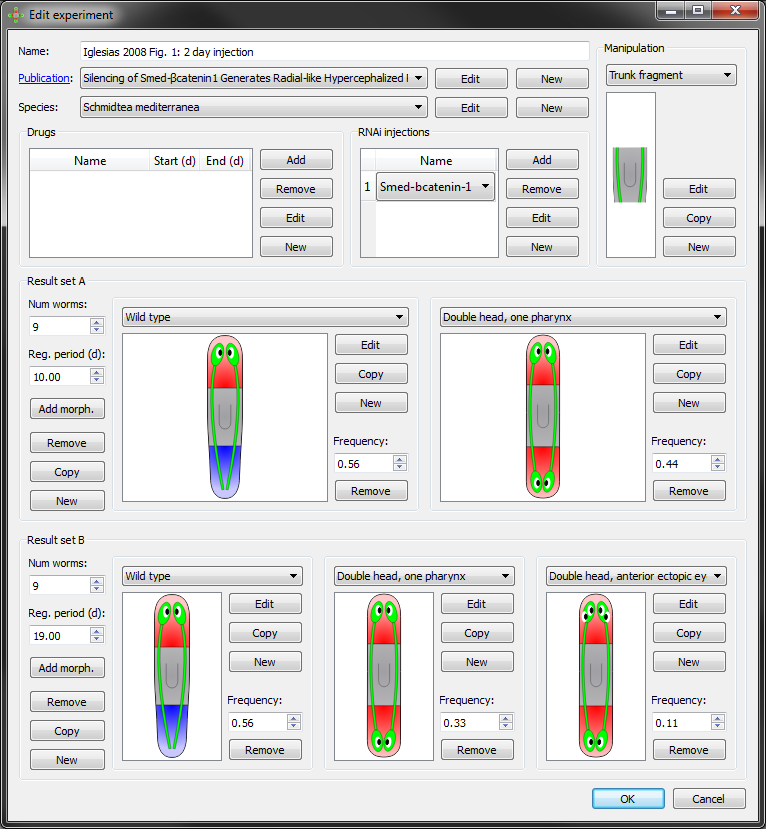
In addition to the raw information stored in the databases, we develop user-friendly graphical interfaces that permit any scientist to access and mine these resources. We create applications for the visualization and optimized search of specific data. For example, all morphological outcomes reported in the literature resulting from certain pharmacological or genetic intervention can be listed and visualized in the application in seconds. Furthermore, these applications can be used for easily formalizing new experiments, which can then be shared and included in scientific publications.
Publications
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, A. Wolff, D. Lobo
Development 151 (9): dev202353, 2024.
(Highlighted and interviewed)
mergem: merging, comparing, and translating genome-scale metabolic models using universal identifiers
A. Hari, A. Zarrabi, D. Lobo
NAR Genomics and Bioinformatics 6(1), lqae010, 2024.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, D. Lobo
bioRxiv doi:10.1101/2023.09.15.557968, 2023.
mergem: merging and comparing genome-scale metabolic models using universal identifiers
A. Hari, D. Lobo
bioRxiv doi:10.1101/2022.07.14.499633, 2022.
Formalizing phenotypes of regeneration
D. Lobo
Whole-Body Regeneration
S. Blanchoud, B. Galliot (eds.)
Springer pp. 663-679, 2022.
Computational systems biology of morphogenesis
J.M. Ko, R. Mousavi, D. Lobo
Computational Systems Biology in Medicine and Biotechnology
S. Cortassa, M.A. Aon (eds.)
Springer pp. 343-365, 2021.
Fluxer: a web application to compute, analyze, and visualize genome-scale metabolic flux networks
A. Hari, D. Lobo
Nucleic Acids Research 48, pp. 427-435, 2020.
Curation and annotation of planarian gene expression patterns with segmented reference morphologies
J. Roy, E. Cheung, J. Bhatti, A. Muneem, D. Lobo
Bioinformatics 36(9), pp. 2881-2887, 2020.
Computing a worm: reverse-engineering planarian regeneration
D. Lobo, M. Levin
Advances in Unconventional Computing: Emergence, Complexity and Computation
A. Adamatzky (ed.)
Springer pp. 637-654, 2017.
MoCha: molecular characterization of unknown pathways
D. Lobo, J. Hammelman, M. Levin
Journal of Computational Biology 23(4): 291-297, 2016.
A dynamic architecture of life
B.P. Rubin, J. Brockes, B. Galliot, U. Grossniklaus, D. Lobo, M. Mainardi, M. Mirouze, A. Prochiantz, A. Steger
F1000Research 4:1288, 2015.
Limbform: a functional ontology-based database of limb regeneration experiments
D. Lobo, E.B. Feldman, M. Shah, T.J. Malone, M. Levin
Bioinformatics 30(24), pp. 3598-3600, 2014.
A bioinformatics expert system linking functional data to anatomical outcomes in limb regeneration
D. Lobo, E.B. Feldman, M. Shah, T.J. Malone, M. Levin
Regeneration 1(2), pp. 37-56, 2014.
Planform: an application and database of graph-encoded planarian regenerative experiments
D. Lobo, T.J. Malone, M. Levin
Bioinformatics 29(8), pp. 1098-1100, 2013.
Towards a bioinformatics of patterning: a computational approach to understanding regulative morphogenesis
D. Lobo, T.J. Malone, M. Levin
Biology Open 2(2), pp. 156-169, 2013.
(Selected for the journal cover)
Modeling planarian regeneration: a primer for reverse-engineering the worm
D. Lobo, W.S. Beane, M. Levin
PLoS Computational Biology 8(4): e1002481, 2012.
(Selected for the journal cover)
Graph grammars with string-regulated rewriting
D. Lobo, F.J. Vico, J. Dassow
Theoretical Computer Science 412(43), pp. 6101-6111, 2011.
(Reviewed in Mathematical Reviews, American Mathematical Society, MR2883035)