Development and Regeneration
Regeneration of body parts
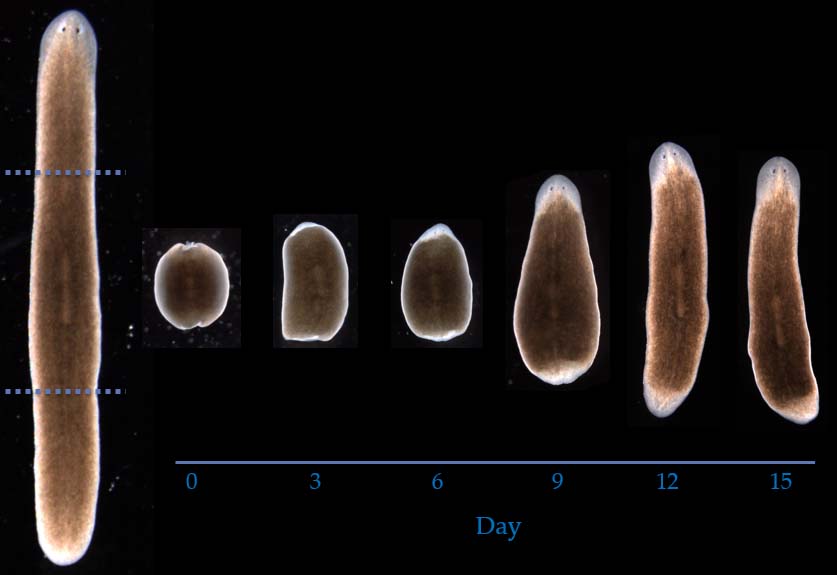
The regeneration of salamander limbs and planarian complete bodies are some of the most striking examples of the restoration of functional and complex structures. Planarian worms, despite a complex body morphology including a true brain, eyes, nervous, digestive, and musculature systems, can regenerate almost any amputated piece, including a new head, trunk, or tail and any missing internal organ. Understanding the regulatory mechanisms controlling these extraordinary abilities will pave the way for revolutionary medical applications. We create automated computational systems that can reverse-engineer dynamic genetic regulatory networks of planarian regeneration from experimental data, propose new hypothesis, and predict new phenotypes and genes to validate at the bench.
Shape and form encodings
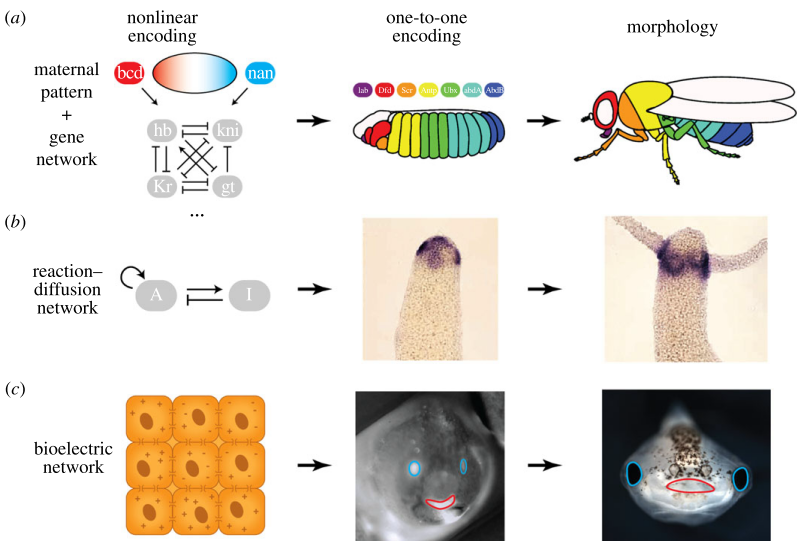
Extraordinary shapes and forms are produced by multicellular organisms during development and regeneration. How and where are these shapes and forms encoded within the organism? We apply computational, information, and mathematical theory to address these complex questions. The theories of inverse problems and linear and non-linear encodings suggest that some highly-plastic regenerative and developmental phenomena can only be explained by certain linear models of information encoding, instead of the assumed classical non-linear mechanisms such as the genome. These findings highlight the need to develop testable models of regulation that combine emergence with a top-down specification of shape.
Evolutionary development
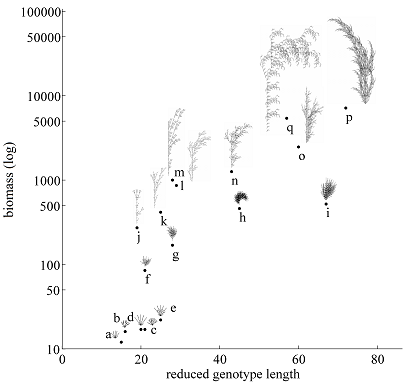
How do the novel and diverse array of morphological forms arise during evolution? We are interested in the properties of evolution and development that allows the emergence of the vast novelty and diversity found in nature. We investigate synthetic systems (ALife) to test the influence of different evo-devo concepts, such as evolutionary pressure, morphological encodings, and open-ended evolution. In addition, we use our discoveries in evolutionary emergence for developing new computational methods applicable to automated creative machines.
Publications
Emergent tissue shapes from the regulatory feedback between morphogens and cell growth
B. Kaity, D. Lobo
bioRxiv doi:10.1101/2025.02.16.638504, 2025.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, A. Wolff, D. Lobo
Development 151 (9): dev202353, 2024.
(Highlighted and interviewed)
Automatic design of gene regulatory mechanisms for spatial pattern formation
R. Mousavi, D. Lobo
NPJ Systems Biology and Applications 10, 35, 2024.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, D. Lobo
bioRxiv doi:10.1101/2023.09.15.557968, 2023.
In situ probe and inhibitory RNA synthesis using streamlined gene cloning with Gibson assembly
A. Wolff, C. Wagner, J. Wolf, D. Lobo
STAR Protocols 3, 101458, 2022.
Formalizing phenotypes of regeneration
D. Lobo
Whole-Body Regeneration
S. Blanchoud, B. Galliot (eds.)
Springer pp. 663-679, 2022.
Inference of dynamic spatial GRN models with multi-GPU evolutionary computation
R. Mousavi, S.H. Konuru, D. Lobo
Briefings in Bioinformatics 22(5), bbab104, 2021.
Computational systems biology of morphogenesis
J.M. Ko, R. Mousavi, D. Lobo
Computational Systems Biology in Medicine and Biotechnology
S. Cortassa, M.A. Aon (eds.)
Springer pp. 343-365, 2021.
Curation and annotation of planarian gene expression patterns with segmented reference morphologies
J. Roy, E. Cheung, J. Bhatti, A. Muneem, D. Lobo
Bioinformatics 36(9), pp. 2881-2887, 2020.
Continuous dynamic modeling of regulated cell adhesion: sorting, intercalation, and involution
J.M. Ko, D. Lobo
Biophysical Journal 117(11), pp. 2166-2179, 2019.
Cross-inhibition of Turing patterns explains the self-organized regulatory mechanism of planarian fission
S. Herath, D. Lobo
Journal of Theoretical Biology 485, 110042, 2019.
Continuous dynamic modeling of regulated cell adhesion
J.M. Ko, D. Lobo
bioRxiv 582429, 2019.
Modeling regenerative processes with Membrane Computing
M. García-Quismondo, M. Levin, D. Lobo
Information Sciences 381, pp. 229-249, 2017.
Computing a worm: reverse-engineering planarian regeneration
D. Lobo, M. Levin
Advances in Unconventional Computing: Emergence, Complexity and Computation
A. Adamatzky (ed.)
Springer pp. 637-654, 2017.
Computational discovery and in vivo validation of hnf4 as a regulatory gene in planarian regeneration
D. Lobo, J. Morokuma, M. Levin
Bioinformatics 32(17), pp. 2681-2685, 2016.
Physiological controls of large-scale patterning in planarian regeneration: a molecular and computational perspective on growth and form
F. Durant, D. Lobo, J. Hammelman, M. Levin
Regeneration 3(2), pp. 78-102, 2016.
(Selected for the journal cover)
Artificial neural networks as models of robustness in development and regeneration: stability of memory during morphological remodeling
J. Hammelman, D. Lobo, M. Levin
Artificial Neural Network Modelling
S. Shanmuganathan, S. Samarasinghe (eds.)
Springer pp. 45-65, 2016.
Gap junctional blockade stochastically induces different species-specific head anatomies in genetically wild-type Girardia dorotocephala flatworms
M. Emmons-Bell, F. Durant, J. Hammelman, N. Bessonov, V. Volpert, J. Morokuma, K. Pinet, D. Adams, A. Pietak, D. Lobo, M. Levin
International Journal of Molecular Sciences 16(11), 27865-27896, 2015.
(Selected for the journal cover)
A dynamic architecture of life
B.P. Rubin, J. Brockes, B. Galliot, U. Grossniklaus, D. Lobo, M. Mainardi, M. Mirouze, A. Prochiantz, A. Steger
F1000Research 4:1288, 2015.
Inferring regulatory networks from experimental morphological phenotypes: a computational method reverse-engineers planarian regeneration
D. Lobo, M. Levin
PLoS Computational Biology 11(6): e1004295, 2015.
Limbform: a functional ontology-based database of limb regeneration experiments
D. Lobo, E.B. Feldman, M. Shah, T.J. Malone, M. Levin
Bioinformatics 30(24), pp. 3598-3600, 2014.
A bioinformatics expert system linking functional data to anatomical outcomes in limb regeneration
D. Lobo, E.B. Feldman, M. Shah, T.J. Malone, M. Levin
Regeneration 1(2), pp. 37-56, 2014.
A linear-encoding model explains the variability of the target morphology in regeneration
D. Lobo, M. Solano, G.A. Bubenik, M. Levin
Journal of the Royal Society Interface 30(24), pp. 3598-3600, 2014.
(Recommended by F1000Prime, Faculty of 1000, 718232471)
Planform: an application and database of graph-encoded planarian regenerative experiments
D. Lobo, T.J. Malone, M. Levin
Bioinformatics 29(8), pp. 1098-1100, 2013.
Towards a bioinformatics of patterning: a computational approach to understanding regulative morphogenesis
D. Lobo, T.J. Malone, M. Levin
Biology Open 2(2), pp. 156-169, 2013.
(Selected for the journal cover)
Behavior-finding: morphogenetic designs shaped by function
D. Lobo, J.D. Fernández, F.J. Vico
Morphogenetic Engineering: Toward Programmable Complex Systems
R. Doursat, H. Sayama, O. Michel (eds.)
Springer-Verlag pp. 441-472, 2013.
Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo
M. Lobikin, B. Chernet, D. Lobo, M. Levin
Physical Biology 9(6): 065002, 2012.
(Selected for the journal cover)
Modeling planarian regeneration: a primer for reverse-engineering the worm
D. Lobo, W.S. Beane, M. Levin
PLoS Computational Biology 8(4): e1002481, 2012.
(Selected for the journal cover)
Emergent diversity in an open-ended evolving virtual community
J.D. Fernández, D. Lobo, G.M. Martín, R. Doursat, F.J. Vico
Artificial Life 18(2), pp. 199-222, 2012.
Graph grammars with string-regulated rewriting
D. Lobo, F.J. Vico, J. Dassow
Theoretical Computer Science 412(43), pp. 6101-6111, 2011.
(Reviewed in Mathematical Reviews, American Mathematical Society, MR2883035)
Evolution of form and function in a model of differentiated multicellular organisms with gene regulatory networks
D. Lobo, F.J. Vico
BioSystems 102(2-3), pp. 112-123, 2010.
Evolutionary development of tensegrity structures
D. Lobo, F.J. Vico
BioSystems 101(3), pp. 167-176, 2010.