Computational Methods
Reverse-engineering regulatory networks
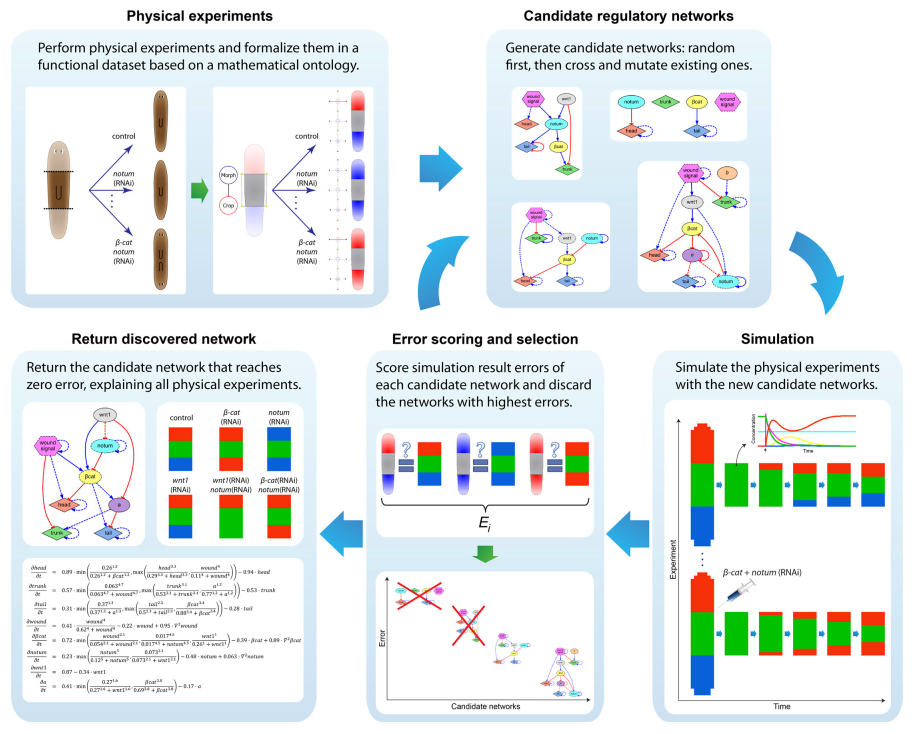
Finding the genetic mechanisms responsible for a given set of resultant phenotypes remains a significant challenge due to both the non-linearity of regulatory networks and the difficulty of testing spatial-temporal models. We investigate computational systems using novel algorithms in machine learning and high-performance computing that can automatically reverse-engineer systems-level dynamic regulatory networks, including all the necessary genes, regulatory interactions, and kinetic parameters, directly from an input dataset of experimental perturbations and their resulting outcomes. Our approach is general enough to be applicable to a diverse set of problems, including complex non-linear signaling networks, genetic and pharmacological experimental perturbations, and spatial-temporal morphological outcomes, paving the way towards automated machine scientists to help us in understanding complex biological regulation.
High performance in silico experiments
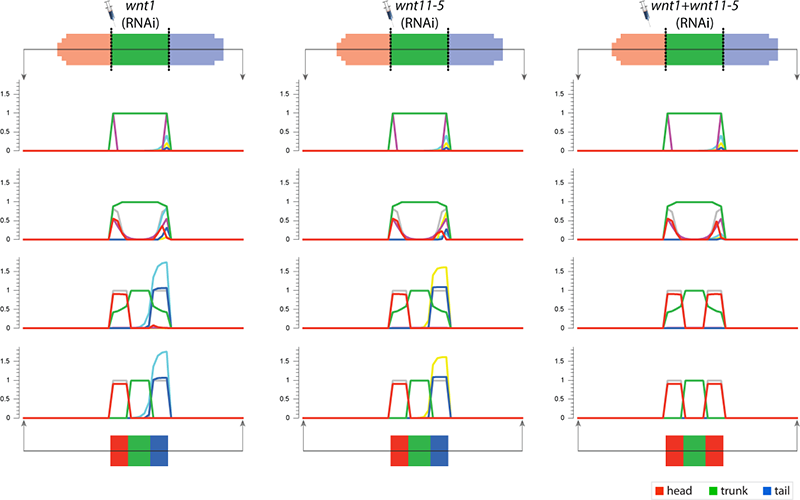
To streamline the discovery of much sought-after systems-level regulatory networks, we develop high-performance simulators capable of performing the same temporal and spatial experiments done at the bench, including genetic knock-downs, pharmacological treatments, and surgical manipulations. In this way, fast in silico experiments can be performed by both human scientists and artificial intelligence machines to discover testable, complete genetic networks that recapitulates the same resultant phenotypes from the experiments in vivo.
Automatic hypothesis generation
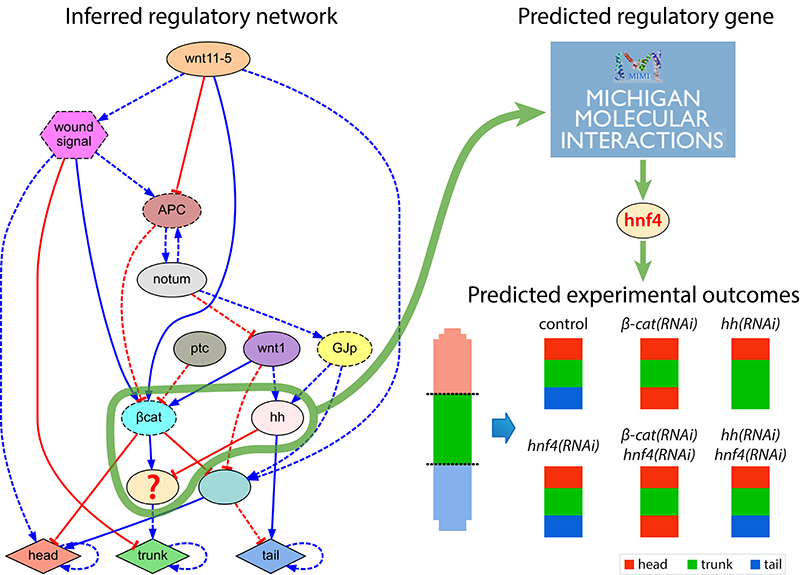
In addition to automatically inferring models from experimental data, we investigate software systems that can also generate novel hypothesis and testable predictions. These systems can automatically discover novel components missing from the input experimental dataset but detected as necessary to explain the resultant phenotypes. Using this approach, we can discover new genes and regulatory interactions and suggest the specific experiments neccesary to validate the new hypothesis at the bench.
Publications
Emergent stable tissue shapes from the regulatory feedback between morphogens and cell growth
B. Kaity, D. Lobo
Journal of Theoretical Biology 620, 112354, 2025.
Predicting the regulatory dynamics of AML disease progression from longitudinal multi-modal clinical data
R. Mousavi, M.K. Mustafa Ali, D. Lobo
Journal of Medical Systems 49, 183, 2025.
FROG analysis ensures the reproducibility of genome scale metabolic models
K. Raman, M. Kratochvil, et al., D. Lobo, et al., R.S. Malik-Sheriff
bioRxiv doi:10.1101/2024.09.24.614797, 2024.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, A. Wolff, D. Lobo
Development 151 (9): dev202353, 2024.
(Highlighted and interviewed)
Automatic design of gene regulatory mechanisms for spatial pattern formation
R. Mousavi, D. Lobo
NPJ Systems Biology and Applications 10, 35, 2024.
mergem: merging, comparing, and translating genome-scale metabolic models using universal identifiers
A. Hari, A. Zarrabi, D. Lobo
NAR Genomics and Bioinformatics 6(1), lqae010, 2024.
Mechanistic regulation of planarian shape during growth and degrowth
J.M. Ko, W. Reginato, D. Lobo
bioRxiv doi:10.1101/2023.09.15.557968, 2023.
mergem: merging and comparing genome-scale metabolic models using universal identifiers
A. Hari, D. Lobo
bioRxiv doi:10.1101/2022.07.14.499633, 2022.
In situ probe and inhibitory RNA synthesis using streamlined gene cloning with Gibson assembly
A. Wolff, C. Wagner, J. Wolf, D. Lobo
STAR Protocols 3, 101458, 2022.
Formalizing phenotypes of regeneration
D. Lobo
Whole-Body Regeneration
S. Blanchoud, B. Galliot (eds.)
Springer pp. 663-679, 2022.
Automatic generation of interactive multidimensional phase portraits
O.O. Ogunsan, D. Lobo
bioRxiv doi:10.1101/2022.02.23.481676, 2022.
Inference of dynamic spatial GRN models with multi-GPU evolutionary computation
R. Mousavi, S.H. Konuru, D. Lobo
Briefings in Bioinformatics 22(5), bbab104, 2021.
Computational systems biology of morphogenesis
J.M. Ko, R. Mousavi, D. Lobo
Computational Systems Biology in Medicine and Biotechnology
S. Cortassa, M.A. Aon (eds.)
Springer pp. 343-365, 2021.
Kinetic modeling of microbial growth, enzyme activity, and gene deletions: an integrated model of β-glucosidase function in Cellvibrio japonicus
J. Hwang, A. Hari, R. Cheng, J.G. Gardner, D. Lobo
Biotechnology and Bioengineering 117, pp. 3876-3890, 2020.
Fluxer: a web application to compute, analyze, and visualize genome-scale metabolic flux networks
A. Hari, D. Lobo
Nucleic Acids Research 48, pp. 427-435, 2020.
Continuous dynamic modeling of regulated cell adhesion: sorting, intercalation, and involution
J.M. Ko, D. Lobo
Biophysical Journal 117(11), pp. 2166-2179, 2019.
Cross-inhibition of Turing patterns explains the self-organized regulatory mechanism of planarian fission
S. Herath, D. Lobo
Journal of Theoretical Biology 485, 110042, 2019.
Continuous dynamic modeling of regulated cell adhesion
J.M. Ko, D. Lobo
bioRxiv 582429, 2019.
Discovering novel phenotypes with automatically inferred dynamic models: a partial melanocyte conversion in Xenopus
D. Lobo, M. Lobikin, M. Levin
Scientific Reports 7, 41339, 2017.
Computing a worm: reverse-engineering planarian regeneration
D. Lobo, M. Levin
Advances in Unconventional Computing: Emergence, Complexity and Computation
A. Adamatzky (ed.)
Springer pp. 637-654, 2017.
Computational discovery and in vivo validation of hnf4 as a regulatory gene in planarian regeneration
D. Lobo, J. Morokuma, M. Levin
Bioinformatics 32(17), pp. 2681-2685, 2016.
Physiological controls of large-scale patterning in planarian regeneration: a molecular and computational perspective on growth and form
F. Durant, D. Lobo, J. Hammelman, M. Levin
Regeneration 3(2), pp. 78-102, 2016.
(Selected for the journal cover)
MoCha: molecular characterization of unknown pathways
D. Lobo, J. Hammelman, M. Levin
Journal of Computational Biology 23(4): 291-297, 2016.
Artificial neural networks as models of robustness in development and regeneration: stability of memory during morphological remodeling
J. Hammelman, D. Lobo, M. Levin
Artificial Neural Network Modelling
S. Shanmuganathan, S. Samarasinghe (eds.)
Springer pp. 45-65, 2016.
A dynamic architecture of life
B.P. Rubin, J. Brockes, B. Galliot, U. Grossniklaus, D. Lobo, M. Mainardi, M. Mirouze, A. Prochiantz, A. Steger
F1000Research 4:1288, 2015.
Serotonergic regulation of melanocyte conversion: A bioelectrically regulated network for stochastic all-or-none hyperpigmentation
M. Lobikin, D. Lobo, D.J. Blackiston, C.J. Martyniuk, E. Tkachenko, M. Levin
Science Signaling 8(397), pp. ra99, 2015.
(Reviewed in a focus paper)
Inferring regulatory networks from experimental morphological phenotypes: a computational method reverse-engineers planarian regeneration
D. Lobo, M. Levin
PLoS Computational Biology 11(6): e1004295, 2015.
Design of a flexible component gathering algorithm for converting cell-based models to graph representations for use in evolutionary search
M. Budnikova, J.W. Habig, D. Lobo, N. Cornia, M. Levin, T. Andersen
BMC Bioinformatics 15:178, 2014.
Behavior-finding: morphogenetic designs shaped by function
D. Lobo, J.D. Fernández, F.J. Vico
Morphogenetic Engineering: Toward Programmable Complex Systems
R. Doursat, H. Sayama, O. Michel (eds.)
Springer-Verlag pp. 441-472, 2013.
Resting potential, oncogene-induced tumorigenesis, and metastasis: the bioelectric basis of cancer in vivo
M. Lobikin, B. Chernet, D. Lobo, M. Levin
Physical Biology 9(6): 065002, 2012.
(Selected for the journal cover)
Modeling planarian regeneration: a primer for reverse-engineering the worm
D. Lobo, W.S. Beane, M. Levin
PLoS Computational Biology 8(4): e1002481, 2012.
(Selected for the journal cover)
Evolution of form and function in a model of differentiated multicellular organisms with gene regulatory networks
D. Lobo, F.J. Vico
BioSystems 102(2-3), pp. 112-123, 2010.
Evolutionary development of tensegrity structures
D. Lobo, F.J. Vico
BioSystems 101(3), pp. 167-176, 2010.
Reconfiguration algorithms for robotically manipulatable structures
D. Lobo, D.A. Hjelle, H. Lipson
Proceedings of the ASME/IFToMM Intern. Conf. on Reconfigurable Mechanisms and Robots
J.S. Dai, M. Zoppi, X. Kong (eds.)
ReMAR2009 pp. 13-22, London, UK, 2009.
Robust off- and online separation of intracellularly recorded up and down cortical states
Y. Seamari, J.A. Narvaez, F.J. Vico, D. Lobo, M.V. Sanchez-Vives
PLoS ONE 2(9): e888, 2007.