Inferring Regulatory Networks from Experimental Morphological Phenotypes:
a Computational Method Reverse-Engineers Planarian Regeneration
Summary
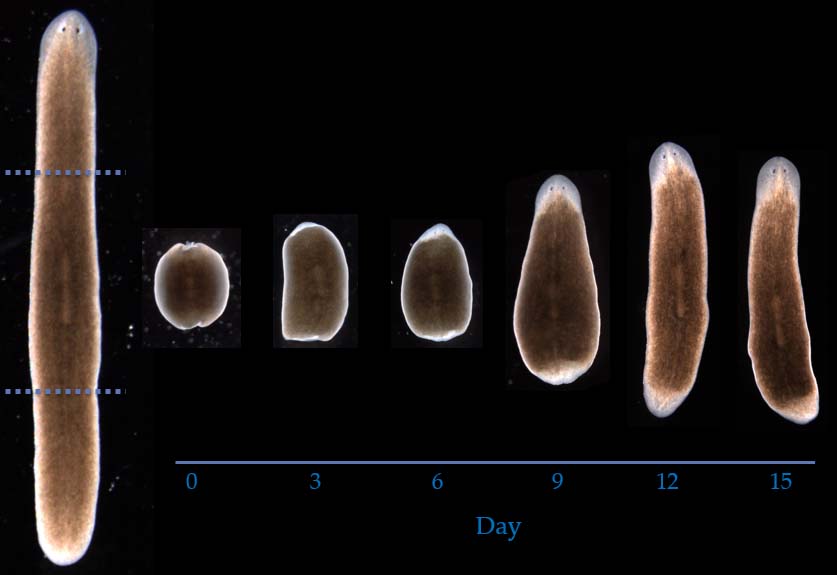
We have reverse-engineered comprehensive dynamic genetic regulatory networks explaining the outstanding capacity of planarian worms to regenerate their head-trunk-tail morphology after the main surgical, genetic, and pharmacological perturbations published in the literature. These are the most comprehensive dynamic regulatory networks discovered to date that can explain the main experiments of head vs. tail regeneration in planarian worms. These systems-biology models are readily predicting novel pathways and genes that will pave the way for our understanding of regeneration.
Software
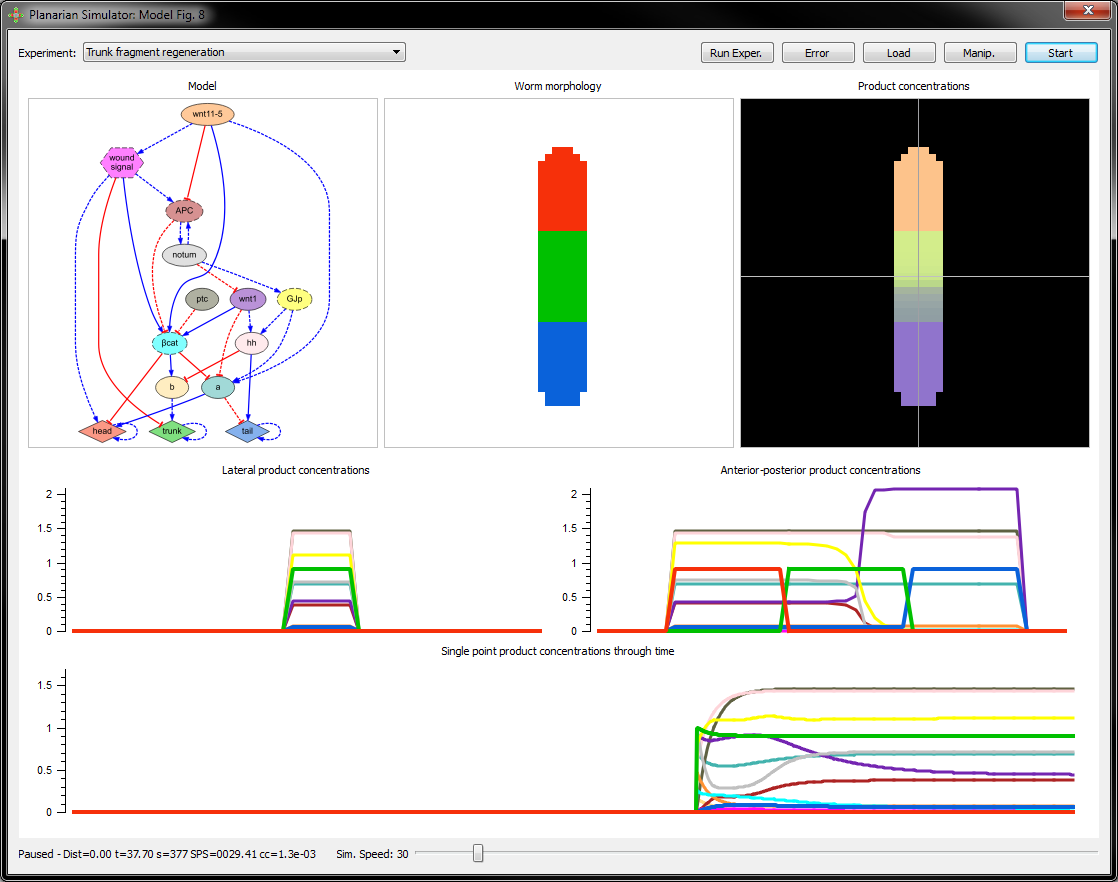
You can freely download the software to simulate in your own computer the automatically reverse-engineered regulatory networks of planarian regeneration from here (Windows 64 bits).
Citation
Inferring regulatory networks from experimental morphological phenotypes: a computational method reverse-engineers planarian regeneration
D. Lobo, M. Levin
PLoS Computational Biology 11(6): e1004295, 2015.
Acknowledgments
This work was supported by NSF grant EF-1124651, NIH grant GM078484, USAMRMC grant W81XWH-10-2-0058, and the Mathers Foundation. Computation used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF grant ACI-1053575, and a cluster computer awarded by Silicon Mechanics.