Streamlined gene cloning with Gibson assembly
Fast cloning for synthesizing in situ riboprobes and RNAi double-stranded RNAs
Overview
We have developed a fast protocol for the synthesis of both riboprobes and double-stranded RNAs for in situ and gene-knockdown experiments. Indeed, in situ hybridization and gene knockdowns are pervasive in the study of developmental biology, but current protocols require time-consuming plasmid restriction digests and inefficient gel purifications. Our novel method uses simultaneous plasmid restriction digestion and Gibson assembly to create vectors that can be used for the synthesis of both single-stranded riboprobes and double-stranded RNAs. This streamlined protocol represents a critical improvement in usability and facilitates the cloning of genes and subsequent WISH, FISH, and RNA interference experiments broadly used in developmental biology.
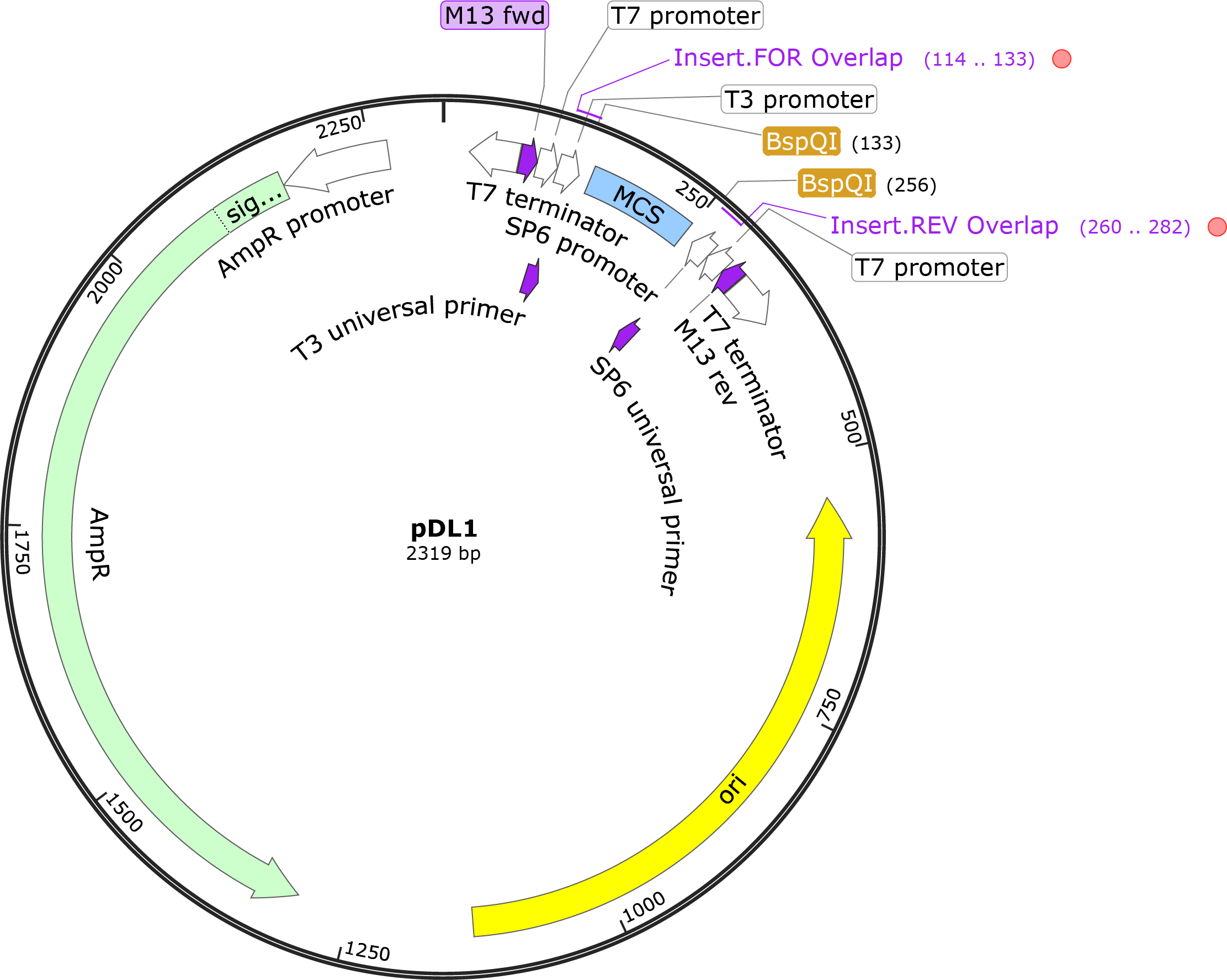
Vector pDL1
We have designed the cloning vector pDL1 for the restriction digest of the vector during the Gibson reaction to insert the gene of interest. pDL1 includes T3 and SP6 promoters for single-stranded RNA synthesis, and two T7 promoters and terminators for dsRNA synthesis. In addition, pDL1 includes a MCS region flanked by BspQI asymmetrical restriction sites to cut the vector during Gibson assembly, M13 forward and reverse universal sequences before the T7 terminators for complete forward and reverse sequencing of the inserted gene, and the AmpR resistance gene. The T3 and SP6 promoter sequences are compatible with their universal primers, as an alternative to the M13 primers for sequencing. The pDL1 plasmid is free to use for nonprofit research and can be purchased through Addgene (cat #182263).
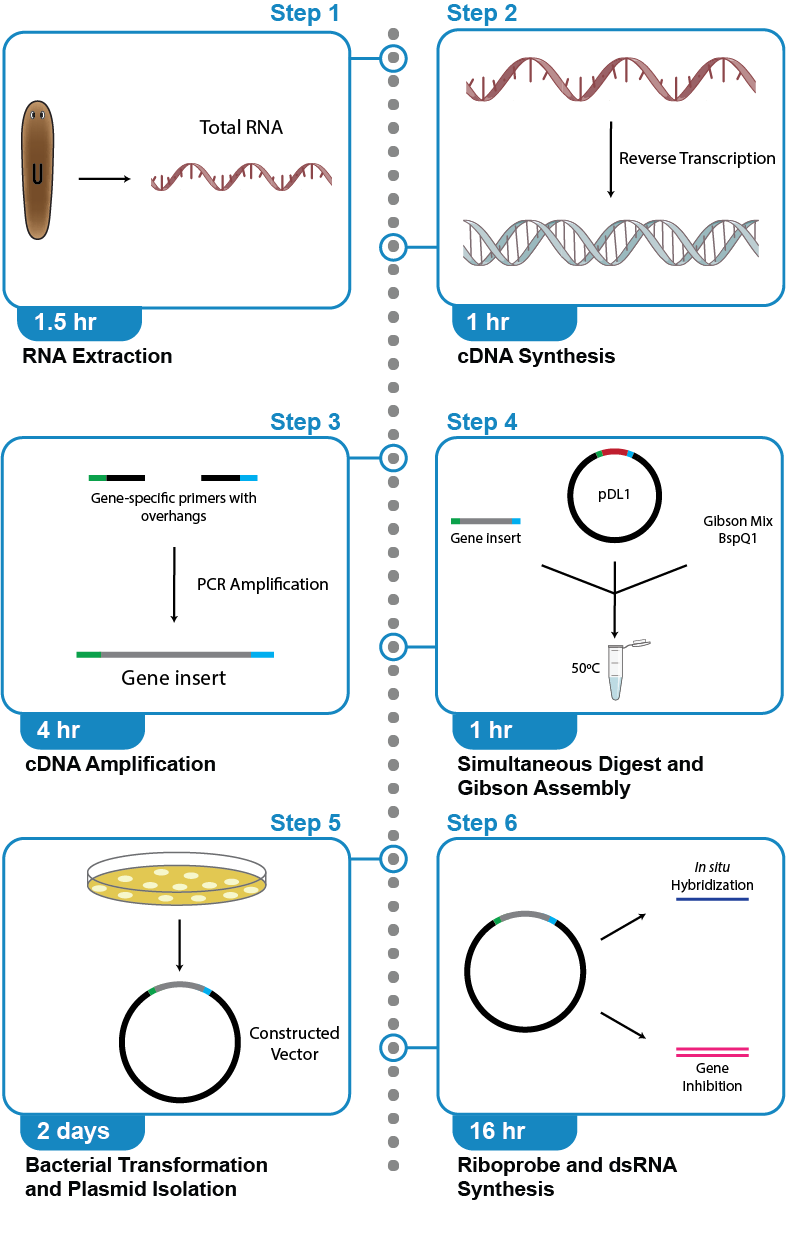
Detailed protocol
We have developed a protocol for gene cloning with pDL1 for synthesizing in situ hybridization riboprobes and double-stranded RNA (dsRNA) for RNA interference (RNAi). We illustrate the protocol with the planarian flatworm Schmidtea mediterranea. However, this protocol can be applied for creating vectors for any organism. Negative and positive controls should be performed for both in situ hybridization riboprobes and dsRNAs. A negative control for riboprobes is synthesizing a probe in the sense direction. A negative control for RNAi can be the sequence for a gene of an unrelated species; we synthesize a dsRNA using the sequence for GFP, which is not native to planarians. We suggest synthesizing a vector containing a sequence like this to serve as a negative control, using a sequence not normally found in your organism. Positive controls for riboprobes and RNAi can be a sequence that has been shown previously to produce a successful in situ hybridization pattern or gene knockdown phenotype.
The detailed protocol is freely-available in this paper published in STAR Protocols (Cell Press). Please use the reference below to cite the protocol in your work.
Citation
In situ probe and inhibitory RNA synthesis using streamlined gene cloning with Gibson assembly
A. Wolff, C. Wagner, J. Wolf, D. Lobo
STAR Protocols 3, 101458, 2022.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM137953. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.